iUTAH Team - Undergraduate iFellows

Rebecca Lee
Brigham Young University
Mentors:
Faculty: Zach Aanderud, BYU
Graduate: Dylan Dastrup, BYU
Neer-Peer: Natasha Griffin, BYU
Research Focus:
Research Focus Area 1
Major:
Environmental Science
Biography:
Rebecca Lee is a junior at Brigham Young University where she is majoring in Environmental Science with a certificate in Spanish. She especially enjoys botany, soil science, and water quality research. After living a year and a half in Nicaragua, she also discovered her love of ecology and travel. In her free time she loves reading, exploring new places, and learning new things. She is excited to contribute to the research that will help improve Utah's water quality and sustainability.
iFellow Presentation:
Identifying and comparing fecal contamination
sources in three Utah watershedse

Presented by: Rebecca Lee
July 2016
Weekly Recap:
Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 | Week 8 | Week 9 | Week 10 | Week 11
Week 1: May 16-20, 2016
What a great start to iUTAH! This week started with orientation and then I began my research at BYU. I was oriented on my project and began to review the literature associated with microbial source tracking and its procedures. We went out and took some GAMUT water samples. It was interesting to see how the river changed along the urban gradient

Visible agricultural pollution in the Provo river in Charleston

Filtering water samples as part of iUTAH GAMUT sampling
Week 2: May 23-27, 2016
After another informative cohort session this week, the week continued with some more literature review so that we could finalize the method for our project. On Wednesday we went and took water samples from two points along the Provo River in order to begin DNA extraction on them. We also learned more about and helped with the GAMUT flow rate measurements and climate stations. Back at the lab, I was able to filter our water samples to capture microbes and extract DNA from those filters. It is exciting to start all of the data collection and research that we have spent the last two weeks preparing for!

DNA extraction
Week 3: May 30-June 3, 2016
Our project deals with microbial source tracking which will allow us to determine non-point contamination sources in Utah's watersheds. This week we continued with DNA extraction and began PCR. We are isolating DNA so that we can make standards to compare future samples to. PCR replicates the DNA we found so that we can analyze it. As a lab, we also went out to Rush Valley, Utah to collect soil samples, do water infiltration tests, and cover surveys.

Taking soil samples at Rush Valley
Week 4: June 6-10, 2016
This week sampling was done at the Logan sites so that we will be able to test them for contamination. Also, I was able to participate in research at Rush Valley. I was able to work on my poster and paper and start putting in words the reason why our research is so important and the methods behind what we are doing.
Week 5: June 13-17, 2016
I definitely feel like we are getting into the swing of things with this past week! After a fun cohort trip on Monday that involved a field trip up the canyon, I was able to continue data collection. There was more DNA extractions to be done, we sampled water at Red Butte Creek and the Provo River, and I was able to preform PCR on the DNA we extracted. I am really enjoying the research I am doing. I love that I can go out and do field work and then go back to the lab to analyze the samples we collected.

Testing river water for E. coli during Monday's cohort session
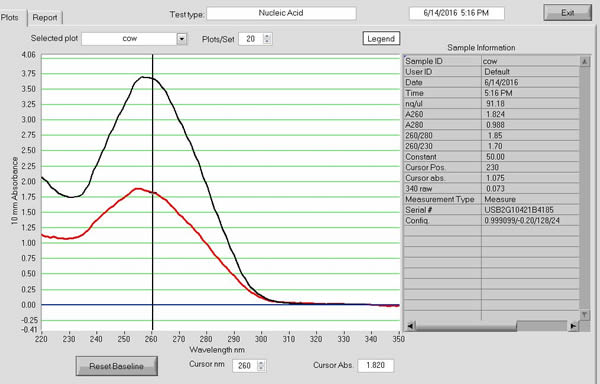
Results from some of my DNA extractions
Week 6: June 20-24, 2016
This video is a fun example of what our water sampling efforts look like! We have taken samples from along the Logan, Provo, and Red Butte watersheds. The filters we use trap bacteria that is in the water. We store the filters in liquid nitrogen so that the microbial community will be the same when we analyze it in the lab as it was at the time we collected it from the river. Once we are back at the lab, we run various procedures so that we can identify if there is fecal contamination from humans, dogs, cows, or other ruminants in the water. With this knowledge, the watersheds will be able to be managed even better. It is interesting how much we can learn from just one simple water sample!
Week 7: June 27-July 1, 2016
This week I extracted DNA from the samples from Logan, Red Butte, and Provo watersheds. The samples are now ready to be analyzed for the specific bacteria we are looking for! We also worked on getting clean PCR results so that we could begin our cloning reactions. We began the cloning process by inserting our PCR results into a circular strand of DNA called a vector. We then put the vector into E. coli cells and cultured them. Doing this allows us to have a quantifiable number to use as a standard by which we can compare our samples from the iUTAH watersheds.

In this step, by putting the cells in a shaking incubator for one hour, the vector is inserted into the E. coli.
Week 8: July 4-8, 2016
This week we worked on cloning and creating standard curves for qPCR. We are planning on starting to do qPCR on our environmental samples in the next few days. I also had the chance to work a lot on my poster for my poster presentation next week. This week the cohort session was Friday and I was able to present a draft of my poster and get very helpful feedback to make it better and help me to better communicate my research.
Week 9: July 11-15, 2016
This week we worked really hard in order to finish running qPCR on our samples from the three different watersheds. This allowed us to relatively quantify how much human, ruminant, and dog contamination was in the samples and see how the amounts of contamination differed along an urban gradient. I also finished my poster and was able to present it at the meeting on Friday. I loved being able to talk about my research with different people and receive feedback. It was also interesting to see the research that everyone else has been doing this summer.
Presenting my poster at the symposium
Week 10: July 18-22, 2016
This week I worked a lot on my oral presentation and on my paper. It is crazy that the iFellows program is almost over. I have enjoyed it so much. Things are wrapping up. It is great that we have been able to relatively quantify fecal contamination. The last thing we are trying to do is make standards for qPCR in order to absolutely quantify the contamination. We are so close to having standards made and have a great protocol for everything except the last two steps. We keep having problems when it comes to having enough plasmid DNA to extract. We first tried making a patch plate, but the colonies didn't grow and then we tried making a liquid culture but the E. coli didn't grow enough in that either. We now are trying a different protocol for the last two steps to see if it works better.

Doing PCR to confirm plasmid insert
Week 11: July 25-29, 2016
I can't believe how fast this summer has gone by! This week I gave my oral presentation and finished up my paper. We also tried to finish making standards, but we won't see if it worked until next week. I am so grateful for the iFellows program and all of the people who made it possible. I have loved being able to learn about microbial source tracking and have my own project. I have loved getting to know and collaborate with the people in my lab and the other iFellows. IUTAH has helped me to discover my love of research and I feel like I have been able to contribute to bettering Utah's water future.
All content provided on this iUTAH Team - Undergraduate iFellows weekly recap is unedited, updated by each participant to provide a review of their progress, and is for informational purposes only.


